Aim 2: Metabolite Annotation & Database Development
Over 95% of compounds found in plant and microbial studies remain structurally and functionally undefined. By developing a comprehensive annotation framework and leveraging large datasets, we aim to expand our understanding of bioactive metabolites that enhance crop resilience.
Research Goals & Approach
1. Developing a Metabolite Annotation Framework
Our team is building the first comprehensive annotation framework focused on plant and microbial metabolites, significantly advancing the current state of metabolomics. Using LC-MS/MS spectral data, we will categorize metabolites into structural, functional, and source-based attributes, extending the ChemFont ontology to include plant- and microbial-specific categories. This FAIR-compliant system integrates aspects from established ontologies such as Gene Ontology and includes tools for structural classification (MSI Levels 1–3), function-based molecular interaction predictions, and source annotation based on phylogenetic and organ-specific data. By addressing the significant underrepresentation of plant and microbial metabolites in spectral databases, this framework will increase annotation rates from <5% to >50%, providing actionable insights into biochemical pathways. Integrated with globally used databases like Global Natural Product Social Molecular Networking (GNPS), Plant Metabolic Network (PMN), and BioAnalytical Resource (BAR), the framework will immediately enhance applications in agriculture, ecological studies, and drug discovery.
2. High-Throughput Data Analysis and Clustering
We employ advanced analytical techniques such as Principal Components Analysis and molecular networking to process the LC-MS/MS datasets generated, uncovering patterns of metabolite changes under stress conditions. By normalizing, filtering, and clustering data using tools like MS-DIAL and GNPS, we will organize bioactive compounds into meaningful clusters that reveal correlations between metabolite profiles and specific stresses. This approach highlights stress-specific biomarkers and conserved metabolic signatures across species, while molecular networks provide insights into metabolite relationships and their broader biological significance. Integrating known libraries such as Library of Active Compounds on Arabidopsis and Library of Pharmacologically Active Compounds will enhance the discovery of structurally similar compounds with immediate agricultural or commercial potential.
3. Database Integration and Visualization
Annotated datasets will be integrated into publicly accessible databases like the GNPS and BAR, ensuring that researchers worldwide can benefit from our findings. These integrations will include enhanced visualization tools that allow users to explore metabolite activity and gene expression across various stresses, tissues, and species. For example, the BAR will adapt its existing frameworks to present metabolite data alongside other omics datasets, offering intuitive interfaces for comparative analysis. By making these tools open-source and user-friendly, we aim to democratize access to high-quality metabolomic data, driving innovation in fields such as crop improvement, biochemistry, and pharmacology.
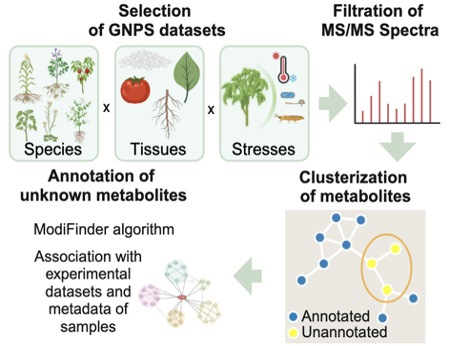
A broad overview of our methodology to annotate unknown metabolites.
Outcomes & Impact
This initiative will result in a comprehensive database of annotated metabolites relevant to crop stress resilience. By increasing the proportion of annotated metabolites from under 5% to over 50%, we create a powerful resource that accelerates pathway discovery and practical applications in breeding and metabolic engineering. Our annotations and visualization tools will support researchers in identifying potential biomarkers, advancing sustainable agriculture, and exploring new genetic targets for crop improvement.
